572g Parameter Estimation of Oscillatory Systems
Oscillatory behavior is common in biology with examples from circadian rhythms [1] to p53-mdm2 DNA damage response [2] to NF-κB oscillations during an immune response [3]. These dynamical behaviors are important and integral in normal biological functions. While there had been considerable modeling and analysis efforts on such systems [4-6], little effort has been spent in reconciling the experimental data with the model output through parameter estimation. Instead, the models were usually built with parameters chosen on an ad-hoc basis (i.e., those that produce oscillations). The purpose of this work is to establish a framework to facilitate data reconciliation between these models and experiments.
Theory and problem formulation:
Two key characteristics of oscillatory behavior are its periodicity and shape. Period is the time interval for one complete oscillation cycle, while shape describes the state trajectory over one cycle. Both contribute to the error between experimental data and model prediction, which is in contrast to standard non-oscillatory systems that only possess shape characteristic and no period. Additional difficulty can arise in oscillatory systems for which the parametric sensitivities and consequently the gradient of objective function in the standard parameter estimation (sum of squares of error), grow unbounded over time [7]. This problem mainly stems from the accumulation of phase differences due to periodicity mismatch.
In this work, we convert time series data to phase-based data by normalization with the periodicity. This serves to decouple the period effect from the shape (contained in the phase-based data). In addition, mismatch in the initial condition between the (noisy) data and model simulation can also contribute to a phase difference. Hence, the initial condition will also be fitted to experimental data.
Methodology:
Based on Maximum Likelihood Estimation [8] under Gaussian error assumption, the objective function is constructed from 2 contributions: period mismatch and shape error. In the parameter estimation, the solution space contains both oscillatory and non-oscillatory solutions. Among the oscillating solutions, only sustained oscillations (i.e., limit cycles) are feasible. Hence, a finite Fourier transform is first used to separate out oscillating solutions when calculating the objective function. Thereafter, oscillation crest comparisons are used to pick out solutions with sustained oscillations. Finally, periods are estimated using the time difference between points of the same phase on each oscillating cycle. These points are chosen to have the steepest gradient to maximize accuracy. Finally, a stochastic optimizer Differential Evolution [9] is used to search for the optimum solution with the lowest objective function value.
Case Studies:
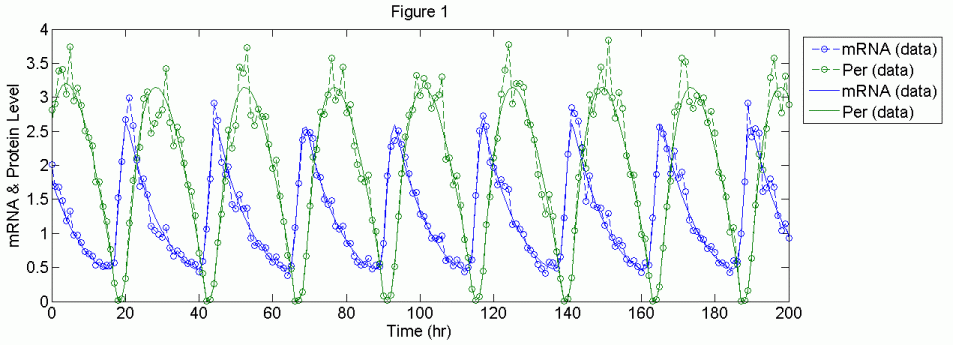
To illustrate this framework’s effectiveness, in-silico data corrupted with 10-20% white noise is generated using published circadian rhythm models for parameter estimation. The use of in-silico data eliminates the issue of plant-model structure mismatch, which allows us to judge the efficacy of the framework in reducing parametric uncertainty. Although the models used are limit cycle models of circadian rhythms [4, 10], the methodology described above has general applicability to any oscillatory systems. The application to both case studies gave solutions with good agreement to experimental data. Figure 1 shows the data and model simulation from a case study [10]. However, some parameter estimates deviate from their true values by as much as 50%. Calculation of the Fisher Information Matrix [8] showed that these parameters have large variances and were practically unidentifiable based on 95% confidence interval. In other words, these calculations imply that the oscillatory behavior is not sensitive to these parameters, which is the main reason for their poor identifiability from experimental data.
References:
[1] J. C. Dunlap. Molecular bases for circadian clocks. Cell, 96:271-290, 1999.
[2] N. Geva-Zatorsky, N. Rosenfeld, S. Itzkovitz, R. Milo, A. Sigal, E. Dekel, T. Yarnitzky, Y. Liron, P. Polak, G. Lahav and U. Alon. Oscillations and variability in the p53 system. Mocular Systems Biology, 2006.0033, 2006.
[3] A. Hoffmann, A. Levchenko, M. L. Scott and D. Baltimore The IkB-NF-kB signaling module, temporal control and selective gene activation. Science, 298:1241-1245, 2002.
[4] A. Goldbeter. A model for circadian oscillations in the Drosophila period protein (PER). Proc. R. Soc. Lond. B, 261:319-324, 1995.
[5] R. L. Bar-or, R. Maya, L. A. Segel, U. Alon, A. J. Levine and M. Oren. Generation of oscillations by the p53-Mdm2 feedback loop: A theoretical and experimental study. Proc. Nat. Acad. Sci., 97:21:11250-11255, 2000.
[6] N. A. M. Monk. Oscillatory expression of Hes1, p53, and NF-kB driven by transcriptional time delays. Current Biology, 13:1409:1413, 2003.
[7] R. Gunawan and F. J. Doyle III. Isochron-based phase response analysis of circadian rhythm. Biophysical Journal, 91:2131-2141, 2006.
[8] Y. Bard. Nonlinear parameter estimation. Academic Press, New York. 1974.
[9] K. V. Price, R. M. Storn and J. A. Lampinen. Differential Evolution: A practical approach to global optimization. Springer, Amsterdam. 2005.
[10] J. J. Tyson, C. I. Hong, C. D. Thron and B. Novak. A simple model of circadian rhythm based on dimerization and proteolysis of PER and TIM. Biophysical Journal, 77:5:2411-2417, 1999.