313c High-Resolution Homology Modeling of Antibody Fv Regions and Application to Antibody-Antigen Docking
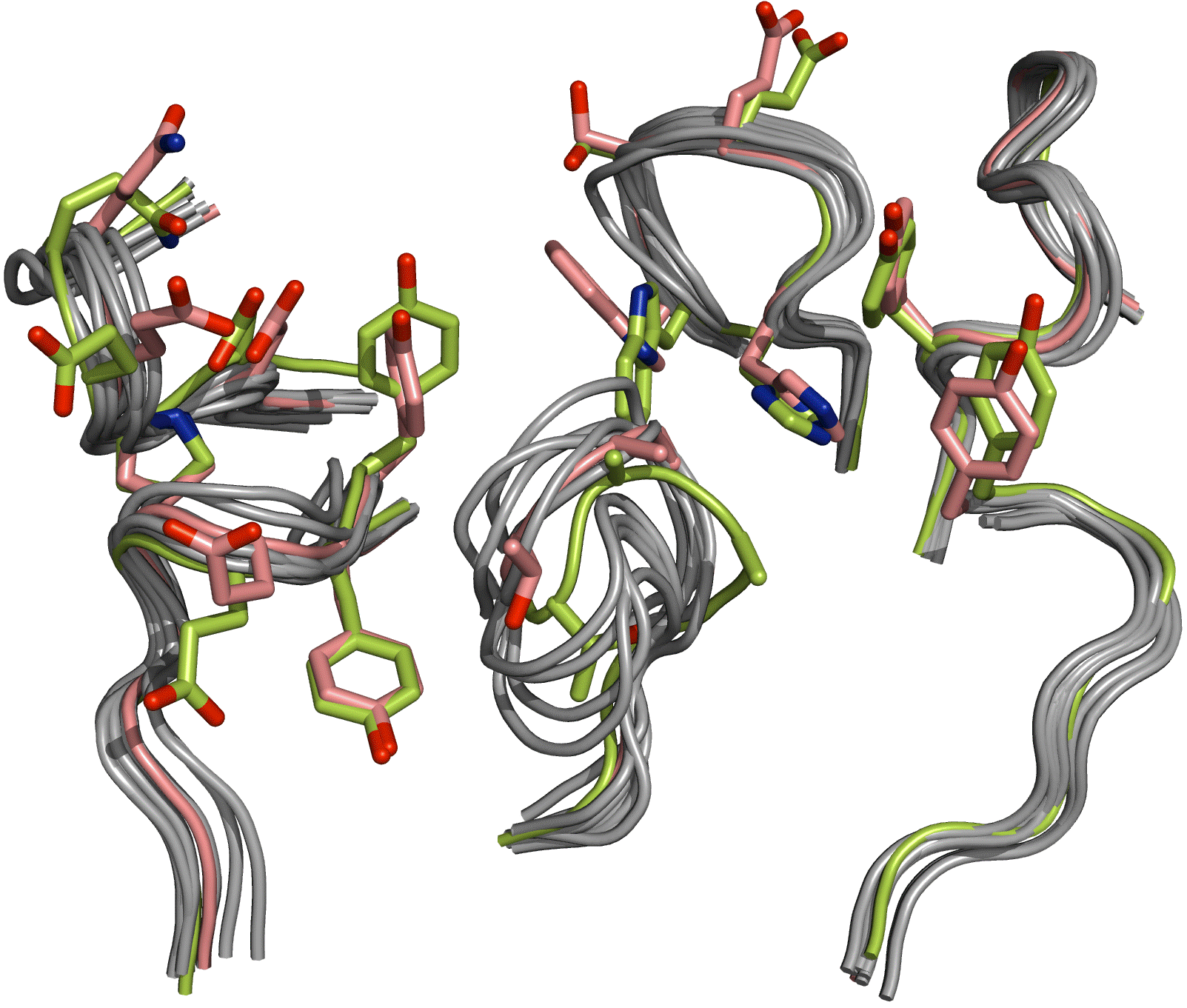
High-resolution homology models are useful in structure-based protein engineering applications when a crystallographic structure is unavailable. Therapeutic antibodies are powerful tools for the prevention and treatment of human and infectious disease because of their high affinity and specificity for target antigens. Furthermore, the existence of many known antibody structures provides a testing platform for high resolution homology modeling tools. Here, we report the development and implementation of RosettaAntibody, a protocol for homology modeling of antibody variable regions. The protocol combines comparative modeling of canonical complementarity determining region (CDR) loop conformations and de novo loop modeling of CDR H3 conformation with simultaneous optimization of VL-VH rigid-body orientation and CDR backbone and side-chain conformations. The protocol was tested on a benchmark of 34 antibody crystal structures. The median root-mean-square-deviation (rmsd) of the antigen binding pocket comprised of all the CDR residues was 1.3 Å with 80% of the targets having an rmsd lower than 2.0 Å. The median rmsd of the final CDR H3 loop prediction was 2.0 Å, and 70% of the loop predictions had an rmsd lower than 2.7 Å. When the set of ten top-scoring antibody homology models are used in local ensemble docking to antigen, a moderate to high accuracy docking prediction was achieved in eight of fifteen targets. This large-scale antibody-antigen docking study using homology models provides insight into the “functional accuracy” of these structural models towards protein engineering applications. The models promise to widen the applicable targets for practical structure-based antibody engineering tasks such as predicting structural changes due to antigen binding, rational design of CDR loops, predicting the structural consequences of antibody humanization including those related to the change in the VL-VH orientation, dissecting structural aspects relevant to antibody cross-reactivity and specificity, and improving antibody affinity. Figure: The antigen binding surface of the ten top-scoring homology models superposed on the antibody crystal structure for D3H44 Fab (1jpt). The native conformation is colored yellow-green and the low-rmsd model is colored salmon. Side chains are shown for the native and low-rmsd structures for CDR residues that contact the antigen in the co-crystal structure.
Web Page: antibody.graylab.jhu.edu/