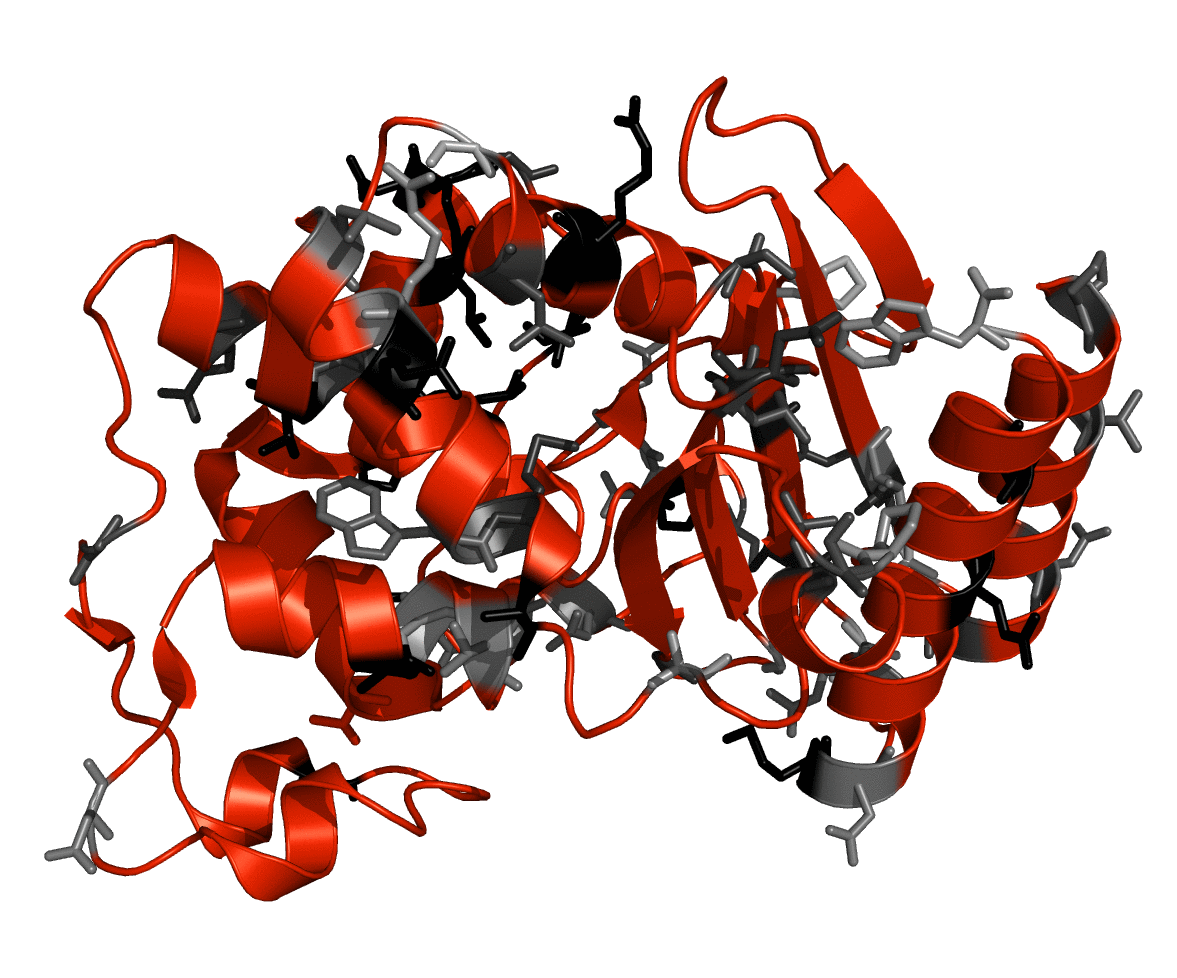504e Computational Predictions of the Enzymatic Activity of Single Deletion Mutants
Single amino acid mutations, (substitutions, insertions, or deletions) in proteins can vary in effect fron negligible to complete loss of function but such effects are not generally predictable. It is generally thought that a deletion is more disruptive to the structure, and thereby the function, of a protein since the gap created must be closed in the folded conformation. Such deletions are widely observed in nature and are often detrimental to the activity and stability of the enzymes, resulting in diseases such as cancer, cystic fibrosis, or osteogenesis imperfecta. Using a set of 50 amino acid deletions from TEM-1 beta-lactamase (Simm et al., FEBS Letters, 581:21, 2007 3904,) we have developed an algorithm to model the perturbed structure of deletion mutants and predict their activity. Preliminary results show that metrics such as distortion of the active site, formation of hydrogen bonding networks, and steric clashes between the ligand and the mutant protein can be used to predict the activity in the mutant proteins. We will further attempt to distinguish mutants which alter substrate concentration required for catalysis (Km) and substrate turnover (kcat). The ability to predict the activity of enzyme mutants has implications for engineering new proteins and understanding the molecular basis of disease. Figure: TEM-1 beta-lactamase with deletion points from the Simm dataset shown in gray. Residues that were deleted are shown in shades of gray representing the amount of activity observed experimentally; no activity (black), wild-type activity (light gray)